REAL Space for novel antibitoic discovery
In an era of escalating antibiotic resistance, there is a pressing need for innovative strategies to develop novel antibiotics. Gram-negative bacteria, characterized by their robust dual-membrane, are intrinsically resistant to a wide range of antibiotics and can readily develop new resistances. Members of this bacterial class comprise numerous pathogenic organisms, including the primary cause of gastric cancer, Helicobacter pylori. In this study, we used the Giga-sized collection of theoretical molecules inside Enamine’s REAL Space to identify inhibitors for H. pylori glutamate racemase. These compounds displayed a diverse range of activity in preventing H. pylori growth, with our most potent hits capable of selective full growth inhibition for metronidazole and clarithromycin resistant H. pylori strains. Alongside the introduction of a novel antibiotic class for this carcinogenic pathogen, our unique implementation of REAL Space holds great promise for Gram-negative antibiotic development as a whole.
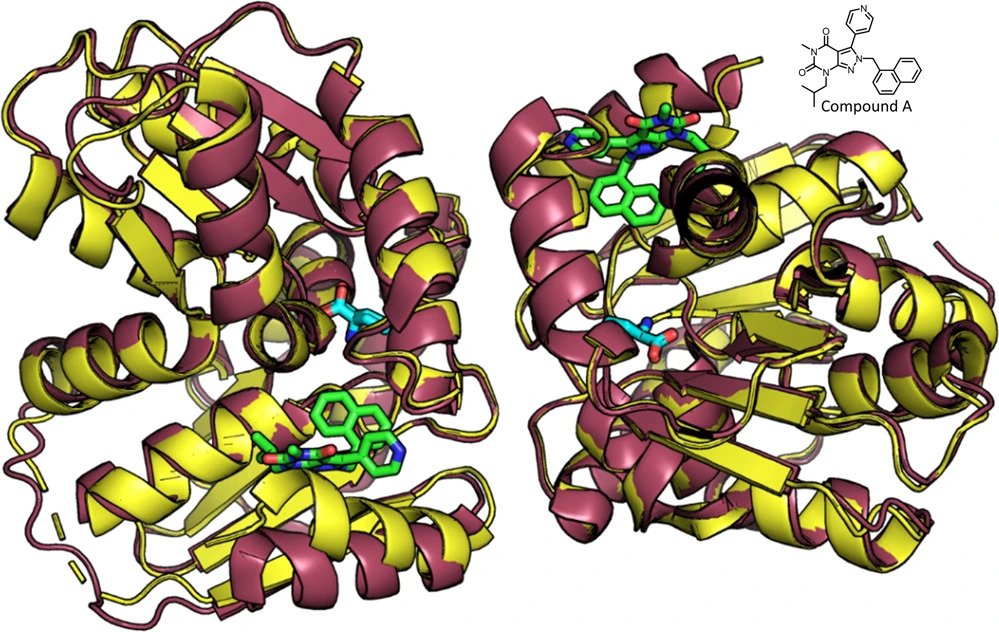
Allosteric Tuning of Caspase-7: Establishing the Nexus of Structure and Catalytic Power
Caspase-7 (C7), a cysteine protease involved in apoptosis, is a valuable drug target for its role in human diseases (e. g., Parkinson's, Alzheimer's, sepsis). The C7 allosteric site has great potential for small-molecule targeting, but numerous drug discovery efforts have identified precious few allosteric inhibitors. Here we present the first selective, drug-like inhibitor of C7 along with several other improved inhibitors based on our previous fragment hit. We also provide a rational basis for the impact of allosteric binding on the C7 catalytic cycle by using an integrated approach including X-ray crystallography, stopped-flow kinetics, and molecular dynamics simulations. Our findings suggest allosteric binding disrupts C7 pre-acylation by neutralization of the catalytic dyad, displacement of substrate from the oxyanion hole, and altered dynamics of substrate binding loops. This work advances drug targeting efforts and bolsters our understanding of allosteric structure–activity relationships (ASARs).
Decrypting a cryptic allosteric pocket in H. pylori glutamate racemase
One of our greatest challenges in drug design is targeting cryptic allosteric pockets in enzyme targets. Drug leads that do bind to these cryptic pockets are often discovered during HTS campaigns, and the mechanisms of action are rarely understood. Nevertheless, it is often the case that the allosteric pocket provides the best option for drug development against a given target. In the current studies we present a successful way forward in rationally exploiting the cryptic allosteric pocket of H. pylori glutamate racemase, an essential enzyme in this pathogen’s life cycle. A wide range of computational and experimental methods are employed in a workflow leading to the discovery of a series of natural product allosteric inhibitors which occupy the allosteric pocket of this essential racemase. The confluence of these studies reveals a fascinating source of the allosteric inhibition, which centers on the abolition of essential monomer-monomer coupled motion networks.
https://doi.org/10.1038/s42004-021-00605-z